
| TerSyn Pathway : Menthol Biosynthesis |
Reaction: EC 1.1.1.208 : (+)-neomenthol + NADP+ = (-)-menthone + NADPH + H+
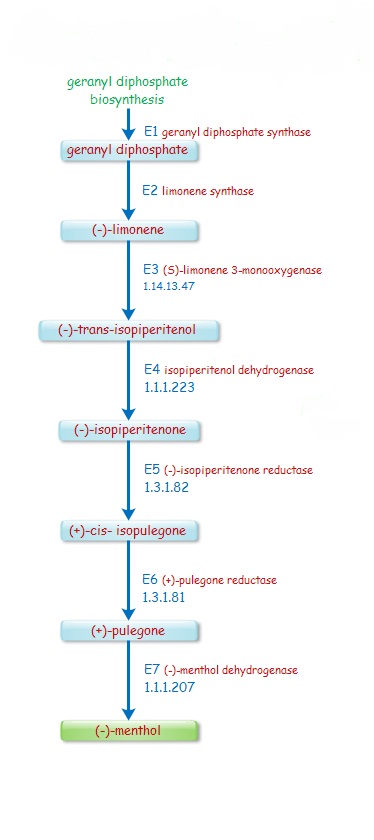
Reaction: EC 1.1.1.207 : (-)-menthol + NADP+ = (-)-menthone + NADPH + H+
Reaction: EC 1.14.13.104 : (+)-pulegone + NADPH + H+ + O2 = (+)-menthofuran + NADP+ + H2O
Reaction: EC 1.3.1.81: (1) (-)-menthone + NADP+ = (+)-pulegone + NADPH + H+
(2) (+)-isomenthone + NADP+ = (+)-pulegone + NADPH + H+ Reaction: EC 1.3.1.82 : (+)-cis-isopulegone + NADP+ = (-)-isopiperitenone + NADPH + H+
Reaction: EC 1.1.1.223 : (-)-trans-isopiperitenol + NAD+ = (-)-isopiperitenone + NADH + H+
Reaction: EC 1.14.13.47: (S)-limonene + NADPH + H+ + O2 = (-)-trans-isopiperitenol + NADP+ + H2O
Reaction: EC 4.2.3.16 : geranyl diphosphate = (S)-limonene + diphosphate |
Menthol biosynthesis pathway mapping The Menthol Biosynthesis Pathway; Mapping Table below shows the complete pathway enzymes responsible for synthesis of Menthol in Mentha Species. The first row shows the enzymes (E1,E2,E3,E4,E5,E6,E7) sequentially as shown in the above figure for menthol biosynthesis; below it shows the step by step mapping of each enzyme with different plants and represent the orthology with Mentha. In the table [YES] represent the presence of the particular ortholog enzyme with its accession number and %Identity. Here [NO] represent the absence of enzyme in that plant i.e it does not show any orthology with particular enzyme. . ....more. |
| S.NO | E1 (Geranyl diphosphate synthase ) (AAF08793.1) (%identity) |
E2 (limonene synthase) |
E3 (limonene 3- mono-oxygenease) [Q6IV13] |
E4 (isopiperitenol dehydrogenase) [Q5C9I9] |
E5 (isopiperitenol reductase) [Q6WAU1]
|
E6 (pulegone reductase) |
E7 ( menthol dehydrogenase) [Q5CAF4]
|
MENTHOL Synthesise |
|||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
1 |
Mentha x piperita
|
YES
|
YES
|
YES
|
YES
|
YES
|
YES
|
|
|||||||
2 |
Mentha canadensis [ABR15420.1/98] |
YES
|
YES
|
YES
|
YES
|
YES
|
YES
|
||||||||
3 |
Ricinus communis [XP_002525096.1/73] |
YES
|
YES
|
YES
|
YES
|
YES
|
YES
|
||||||||
4 |
Vitis vinifera [XP_002273789.1/ 69] |
YES
|
NO |
YES
|
YES
|
YES
|
YES
|
||||||||
5 |
Glycine max [XP_003547995.1/ 69] |
NO |
YES
|
YES
|
YES
|
YES
|
YES
|
||||||||
6 |
Prunus persica [EMJ05575.1/69] |
NO |
YES
|
YES
|
YES
|
YES
|
YES
|
||||||||
7 |
Solanum tuberosum [XP_006363135.1/68] |
NO |
YES
|
YES
|
YES
|
YES
|
YES
|
||||||||
8 |
Citrus sinensis [XP_006466719.1/ 65] |
NO |
YES
|
YES
|
YES
|
YES
|
YES
|
||||||||
9 |
Solanum lycopersicum [NP_001234087.1/ 67] |
NO |
YES
|
YES
|
YES
|
YES
|
YES
|
||||||||
10 |
Phaseolus vulgaris [ESW28009.1/ 68] |
NO |
YES
|
YES
|
YES
|
YES
|
YES
|
||||||||
11 |
Medicago truncatula [XP_003629488.1/ 71] |
NO |
YES
|
YES
|
YES
|
NO |
YES
|
||||||||
12 |
Citrus clementina [XP_006425738.1/ 64] |
NO |
YES
|
YES
|
YES
|
NO |
YES
|
||||||||
13 |
Theobroma cacao [EOX91186.1/ 65] |
NO |
N0 |
YES
|
YES
|
||||||||||
14 |
Eutrema salsugineum [XP_006411959.1/ 75] |
NO |
NO |
YES
|
YES
|
||||||||||
15 |
Arabidopsis lyrata [XP_002869021.1/ 76] |
NO |
NO |
NO |
|||||||||||
16 |
Arabidopsis thaliana [NP_195399.1/ 68] |
NO |
NO |
YES
|
NO |
||||||||||
17 |
Populus trichocarpa [XP_002310785.1/ 74] |
NO |
NO |
YES
|
|||||||||||
18 |
Capsella rubella [XP_006282650.1/ 76] |
NO |
NO |
YES
|
NO |
||||||||||
19 |
Cicer arietinum [XP_004509239.1/ 70] |
NO |
YES
|
NO |
NO |
||||||||||
20 |
Fragaria vesca subsp. vesca [XP_004300598.1/ 70] |
NO |
YES
|
YES
|
NO |
NO |
|||||||||
21 |
Amborella trichopoda [ERN17565.1/ 71] |
NO |
NO |
YES
|
NO |
||||||||||
22 |
Capsicum annuum [P80042.1/ 64] |
NO |
YES
|
NO |
NO |
||||||||||
23 |
Salvia miltiorrhiza [AEZ55681.1/ 77] |
YES
|
YES
|
NO |
NO |
NO |
NO |
||||||||
24 |
Adonis aestivalis var. palaestina [AAV74396.1/ 76] |
NO |
NO |
YES
|
NO |
NO |
NO |
||||||||
25 |
Catharanthus roseus [AGL91645.1/ 69] |
NO |
YES
|
NO |
NO |
NO |
NO |
||||||||
26 |
Cucumis melo [AGN48013.1/ 78] |
NO |
NO |
YES
|
NO |
NO |
NO |
References |
|
| 1. | "(-)-Menthol biosynthesis and molecular genetics". Croteau RB1, Davis EM, Ringer KL, Wildung MR. |
| 2. | "Enhanced specificity of mint geranyl pyrophosphate synthase by modifying the R-loop interactions." Hsieh F.L., Chang T.H., Ko T.P., Wang A.H. J. Mol. Biol. 404:859-873(2010) [PubMed] [Europe PMC] [Abstract] |
| 3. | "Regiospecific cytochrome P450 limonene hydroxylases from mint (Mentha) species: cDNA isolation, characterization, and functional expression of (-)-4S-limonene-3-hydroxylase and (-)-4S-limonene-6-hydroxylase."\Lupien99: Lupien S, Karp F, Wildung M, Croteau R (1999). Arch Biochem Biophys 368(1);181-92. PMID: 10415126 |
| 4. | "Geranyl diphosphate synthase: cloning, expression, and characterization of this prenyltransferase as a heterodimer." Burke C.C., Wildung M.R., Croteau R. Proc. Natl. Acad. Sci. U.S.A. 96:13062-13067(1999) [PubMed] [Europe PMC] [Abstract] |